Education Experience
| 2001-2005 | Bachelor Applied Chemistry | Peking University |
| 2005-2009 | Ph. D. Analytical Chemistry | University of Wisconsin-Madison |
Professional Experience
| 2010-2016 | Associate Professor | Tianjin Medical University |
| 2016-2019 | Professor | Tianjin Medical University |
| 2019-2019 | Professor | Tianjin University |
Research Area
Honors and Awards
| Tianjin Young Elite Scientists Sponsorship Program |
| Tianjin “131” Innovative Talents |
| Tianjin Youth Talents for Science and Technology |
Patents
Highlighted Publications
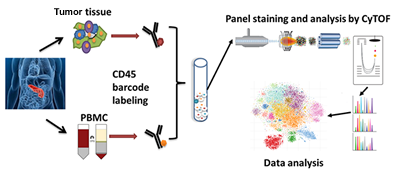
| Wang H#, Chen L#, Qi L, Jiang N, Zhang Z, Guo H, Song T, Li J, Li H, Zhang N*, Chen R*. A single-cell atlas of tumor-infiltrating immune cells in pancreatic ductal adenocarcinoma. Mol Cell Proteomics. 2022 Jun 17;21(8):100258. |
| Wang H, Shi X, Gao Y, Zhang X, Zhao H, Wang L, Zhang X, Chen R*. Polystyrene nanoplastics induce profound metabolic shift in human cells as revealed by integrated proteomic and metabolomic analysis. Environ Int. 2022 Jun 15;166:107349 |
| Xu J, Guan X, Jia X, Li H, Chen R*, Lu Y*. In-depth profiling and quantification of the lysine acetylome in hepatocellular carcinoma with a trapped ion mobility mass spectrometer. Mol Cell Proteomics. 2022 Jun 8;21(8):100255. |
| Yang L, Wang H, Lu W, Yang G, Lin Z, Chen R*, Li H*. Quantitative proteomic analysis of oxaliplatin induced peripheral neurotoxicity. J Proteomics. 2022 Jul 10;266:104682. |
| Wang Z, Ma W, Wei J, Lan K, Yan S, Chen R*, Qin G*. High-performance olfactory receptor-derived peptide sensor for trimethylamine detection based on Steglich esterification reaction and native chemical ligation connection. Biosens Bioelectron. 2022;195:113673. |
| Jiang N#, Gao Y#, Xu J, Luo F, Zhang X, Chen R*. A data-independent acquisition (DIA)-based quantification workflow for proteome analysis of 5000 cells. J Pharm Biomed Anal. 2022;216:114795. |
| Wei J#, Zhao Z#, Lan K, Wang Z, Qin G*, Chen R*. Highly sensitive detection of multiple proteins from single cells by MoS2-FET biosensors. Talanta. 2022; 236:122839. |
| Wei J, Zhao Z, Luo F, Lan K, Chen R*, Qin G*. Sensitive and quantitative detection of SARS-CoV-2 antibodies from vaccinated serum by MoS2-field effect transistor. 2D Mater. 2022;9:015030. |
| Zhao D#, Wang C#, Yan S, Chen R*. Advances in the identification of long non-coding RNA binding proteins. Anal Biochem. 2021;639:114520. |
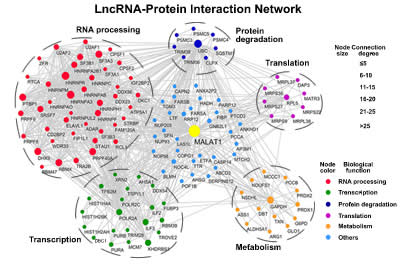
| Wang H, Zhang Y, Guan X, Li X, Zhao Z, Gao Y, Zhang X, Chen R*. An integrated transcriptomics and proteomics analysis implicates lncRNA MALAT1 in the regulation of lipid metabolism. Mol Cell Proteomics. 2021;100141. |
| Yan S, Zhao D, Wang C, Wang H, Guan X, Gao Y, Zhang X, Zhang N*, Chen R*. Characterization of RNA-binding proteins in the cell nucleus and cytoplasm. Anal Chim Acta. 2021;1168:338609. |
| Wang C, Li Y, Yan S, Wang H, Shao X, Xiao M, Yang B, Qin G, Kong R, Chen R*, Zhang N*. Interactome Analysis Reveals that LncRNA HULC Promotes Aerobic Glycolysis through LDHA and PKM2. Nat Commun. 2020;11(1):3162. |
| Dang Y, Jiang N, Wang H, Chen X, Gao Y, Zhang X, Qin G, Li Y*, Chen R*. Proto-oncogene serine/threonine kinase PIM3 promotes cell migration via modulating Rho GTPase signaling. J Proteome Res. 2020;19(3):1298-1309. |
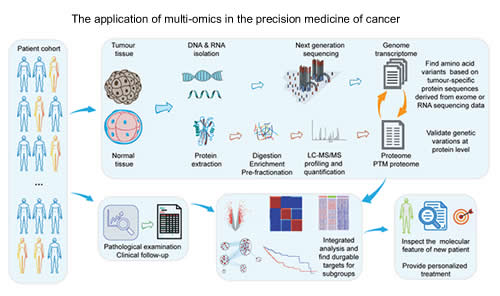
| Wang H, Jiang N, Zhang Y, Chen R*. Clinical proteomics: a driving force for cancer therapeutic target discovery and precision medicine. Cancer Biol Med. 2019;16(4):623-629. |
| Li Y, Zhuang H, Zhang X,Li Y, Liu Y,Yi X, Qin G, Wei W*, Chen R*. Multi-omics integration reveals the landscape of pro-metastasis metabolism in hepatocellular carcinoma. Mol Cell Proteomics. 2018; 17(4):607-618. |
| Wang H, Shao X, He Q, Wang C, Xia L, Yue D, Qin G, Jia C, Chen R*. Quantitative Proteomics Implicates Rictor/mTORC2 in Cell Adhesion. J Proteome Res. 2018 ;17(10):3360-3369 |
| Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, Hou C, Gao H, Zhang X, Jia C, Li L, Li Y*, Zhang N*. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017; 45(17):9947-9959. |
| Qin G, Dang M, Gao H, Wang H, Luo F, Chen R*. Deciphering the protein-protein interaction network regulating hepatocellular carcinoma metastasis. Biochim Biophys Acta. 2017; 1865(9):1114-1122. |
| Chen R*, Xiao M, Gao H, Chen Y, Li Y, Liu Y, Zhang N*. Identification of a novel mitochondrial interacting protein of C1QBP using subcellular fractionation coupled with CoIP-MS. Anal Bioanal Chem. 2016; 408(6):1557-64. |
| Wang Y, Yue D, Xiao M, Qi C, Chen Y, Sun D, Zhang N*, Chen R*.C1QBP negatively regulates the activation of oncoprotein YBX1 in the renal cell carcinoma as revealed by interactomics analysis. J Proteome Res. 2015; 14(2): 804-13. |
| Gao H, Chen Y, Zuo D, Xiao M, Li Y, Guo H, Zhang N, Chen R*. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol Med. 2015; 12(3):246-54. |
| Chen R, Xiao M, Buchberger A, Li L*. Quantitative neuropeptidomics study of the effects of temperature change in the crab Cancer borealis. J Proteome Res. 2014; 13(12):5767-76. |
| Chen R*, Wang Y, Liu Y, Zhang Q, Zhang X, Zhang F, Shieh CH, Yang D, Zhang N*.Quantitative study of the interactome of PKCζ involved in the EGF-induced tumor cell chemotaxis. J Proteome Res. 2013; 12(3):1478-86. |
| Zhang X, Zhang F, Guo L, Wang Y, Zhang P, Wang R, Zhang N*, Chen R*. Interactome analysis reveals that C1QBP (complement component 1, q subcomponent binding protein) is associated with cancer cell chemotaxis and metastasis. Mol Cell Proteomics. 2013; 12(11):3199-209. |